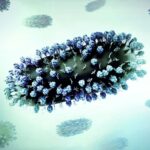
Stanford researchers use single cell genomics analysis (left) to reverse engineer the development of alveolar sacs in the lung (right). Credit: Barbara Treutlein
In a feat of reverse tissue engineering, Stanford researchers have begun to unravel the complex genetic coding that allows embryonic cells to proliferate and transform into all of the specialized cells that perform a myriad of different biological tasks.
A team of interdisciplinary researchers took lung cells from the embryos of mice, choosing samples at different points in the creature’s development cycle. Using the new technique of single-cell genomic analysis, they recorded what genes were active in each cell at each point. Though they studied lung cells, their technique is applicable to any type of cell.
“This lays out a playbook for how to do reverse tissue engineering,” said Stephen Quake, the Lee Otterson Professor in the School of Engineering and leader of the research team.
Researchers used this reverse engineering approach to study the cells in the alveoli – the small balloon-like structures at the tips of the airways. The alveoli serve as docking stations where blood vessels receive oxygen and deliver carbon dioxide.